
Back معزز Arabic Amplificador genètic Catalan Enhancer Czech Enhancer (Genetik) German Ενισχυτής (γενετική) Greek Enhancer Spanish افزایشگر Persian Amplificateur (biologie) French Feabhsaitheoir (bitheolaíocht) Irish Amplificador xenético Galician

- DNA
- Enhancer
- Promoter
- Gene
- Transcription Activator Protein
- Mediator Protein
- RNA Polymerase
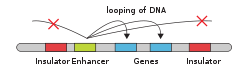
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur.[1][2] These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp (1,000,000 bp) away from the gene, upstream or downstream from the start site.[2][3] There are hundreds of thousands of enhancers in the human genome.[2] They are found in both prokaryotes and eukaryotes.[4] Active enhancers typically get transcribed as enhancer or regulatory non-coding RNA, whose expression levels correlate with mRNA levels of target genes.[5][6]
The first discovery of a eukaryotic enhancer was in the immunoglobulin heavy chain gene in 1983.[7][8][9] This enhancer, located in the large intron, provided an explanation for the transcriptional activation of rearranged Vh gene promoters while unrearranged Vh promoters remained inactive.[10] Lately, enhancers have been shown to be involved in certain medical conditions, for example, myelosuppression.[11] Since 2022, scientists have used artificial intelligence to design synthetic enhancers and applied them in animal systems, first in a cell line,[12] and one year later also in vivo.[13][14]
- ^ Blackwood EM, Kadonaga JT (July 1998). "Going the distance: a current view of enhancer action". Science. 281 (5373): 60–63. Bibcode:1998Sci...281...60.. doi:10.1126/science.281.5373.60. PMID 9679020. S2CID 11666739.
- ^ a b c Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (April 2013). "Enhancers: five essential questions". Nature Reviews. Genetics. 14 (4): 288–295. doi:10.1038/nrg3458. PMC 4445073. PMID 23503198.
- ^ Maston GA, Evans SK, Green MR (2006). "Transcriptional regulatory elements in the human genome". Annual Review of Genomics and Human Genetics. 7: 29–59. doi:10.1146/annurev.genom.7.080505.115623. PMID 16719718. S2CID 12346247.
- ^ Kulaeva OI, Nizovtseva EV, Polikanov YS, Ulianov SV, Studitsky VM (December 2012). "Distant activation of transcription: mechanisms of enhancer action". Molecular and Cellular Biology. 32 (24): 4892–4897. doi:10.1128/MCB.01127-12. PMC 3510544. PMID 23045397.
- ^ Yosefzon Y, David C, Tsukerman A, Pnueli L, Qiao S, Boehm U, et al. (19 September 2017). "An epigenetic switch repressing Tet1 in gonadotropes activates the reproductive axis". Proceedings of the National Academy of Sciences. 114 (38): 10131–10136. doi:10.1073/pnas.1704393114. ISSN 0027-8424. PMC 5617270. PMID 28855337.
- ^ Burren OS, Rubio García A, Javierre BM, Rainbow DB, Cairns J, Cooper NJ, et al. (4 September 2017). "Chromosome contacts in activated T cells identify autoimmune disease candidate genes". Genome Biology. 18 (1): 165. doi:10.1186/s13059-017-1285-0. ISSN 1474-760X. PMC 5584004. PMID 28870212.
- ^ Mercola M, Wang XF, Olsen J, Calame K (August 1983). "Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus". Science. 221 (4611): 663–665. Bibcode:1983Sci...221..663M. doi:10.1126/science.6306772. PMID 6306772.
- ^ Banerji J, Olson L, Schaffner W (July 1983). "A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes". Cell. 33 (3): 729–740. doi:10.1016/0092-8674(83)90015-6. PMID 6409418. S2CID 23981549.
- ^ Gillies SD, Morrison SL, Oi VT, Tonegawa S (July 1983). "A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene". Cell. 33 (3): 717–728. doi:10.1016/0092-8674(83)90014-4. PMID 6409417. S2CID 40313833.
- ^ Hauptman G, Reichert MC, Abdal Rhida MA, Evans TA (December 2022). "Characterization of enhancer fragments in Drosophila robo2". Fly. 16 (1): 312–346. bioRxiv 10.1101/2022.08.01.502399. doi:10.1080/19336934.2022.2126259. PMC 9559326. PMID 36217698.
- ^ Zhigulev A, Norberg Z, Cordier J, Spalinskas R, Bassereh H, Björn N, et al. (March 2024). "Enhancer mutations modulate the severity of chemotherapy-induced myelosuppression". Life Science Alliance. 7 (3): e202302244. doi:10.26508/lsa.202302244. PMC 10796589. PMID 38228368.
- ^ de Almeida BP, Reiter F, Pagani M, Stark A (May 2022). "DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers". Nature Genetics. 54 (5): 613–624. doi:10.1038/s41588-022-01048-5. PMID 35551305.
- ^ de Almeida BP, Schaub C, Pagani M, Secchia S, Furlong EE, Stark A (February 2024). "Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo". Nature. 626 (7997): 207–211. Bibcode:2024Natur.626..207D. doi:10.1038/s41586-023-06905-9. PMC 10830412. PMID 38086418.
- ^ Taskiran II, Spanier KI, Dickmänken H, Kempynck N, Pančíková A, Ekşi EC, et al. (February 2024). "Cell-type-directed design of synthetic enhancers". Nature. 626 (7997): 212–220. Bibcode:2024Natur.626..212T. doi:10.1038/s41586-023-06936-2. PMC 10830415. PMID 38086419.