
Back V-ATPáza Czech V-tüüpi ATPaasid Estonian V-ATPase French ATPase V Galician V형 ATP가수분해효소 Korean V-ATPase Portuguese
| V-ATPase | |
|---|---|
 V-ATPase schematic | |
| Identifiers | |
| Symbol | V-ATPase |
| TCDB | 3.A.2 |
| OPM superfamily | 5 |
| OPM protein | 2bl2 |
| Membranome | 226 |
| V-ATPase, subunit c (Vo) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Membrane-spanning region of the V-type sodium ATPase from Enterococcus hirae. Calculated hydrocarbon boundaries of the lipid bilayer are shown by red and blue dots | |||||||||
| Identifiers | |||||||||
| Symbol | ATP-synt_C | ||||||||
| Pfam | PF00137 | ||||||||
| InterPro | IPR002379 | ||||||||
| PROSITE | PDOC00526 | ||||||||
| SCOP2 | 1aty / SCOPe / SUPFAM | ||||||||
| |||||||||
| V-ATPase, subunit C (V1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of subunit C (vma5p) of the yeast v-atpase | |||||||||
| Identifiers | |||||||||
| Symbol | V-ATPase_C | ||||||||
| Pfam | PF03223 | ||||||||
| InterPro | IPR004907 | ||||||||
| SCOP2 | 1u7l / SCOPe / SUPFAM | ||||||||
| |||||||||
| V-ATPase, subunit I/a | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | V_ATPase_I | ||||||||
| Pfam | PF01496 | ||||||||
| InterPro | IPR002490 | ||||||||
| SCOP2 | 3rrk / SCOPe / SUPFAM | ||||||||
| TCDB | 3.A.2 | ||||||||
| |||||||||
| V-ATPase, subunit E | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | vATP-synt_E | ||||||||
| Pfam | PF01991 | ||||||||
| Pfam clan | CL0255 | ||||||||
| InterPro | IPR002842 | ||||||||
| |||||||||
| V-ATPase, subunit d/d2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of subunit C (yeast subunit d) of v-atpase | |||||||||
| Identifiers | |||||||||
| Symbol | vATP-synt_AC39 | ||||||||
| Pfam | PF01992 | ||||||||
| InterPro | IPR002843 | ||||||||
| SCOP2 | 1r5z / SCOPe / SUPFAM | ||||||||
| |||||||||
| V-ATPase, subunit H, N-terminal | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of the regulatory subunit H of the v-type atpase of saccharomyces cerevisiae | |||||||||
| Identifiers | |||||||||
| Symbol | V-ATPase_H_N | ||||||||
| Pfam | PF03224 | ||||||||
| Pfam clan | CL0020 | ||||||||
| InterPro | IPR004908 | ||||||||
| SCOP2 | 1ho8 / SCOPe / SUPFAM | ||||||||
| |||||||||
| V-ATPase, subunit G | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | V-ATPase_G | ||||||||
| Pfam | PF03179 | ||||||||
| Pfam clan | CL0255 | ||||||||
| InterPro | IPR005124 | ||||||||
| |||||||||
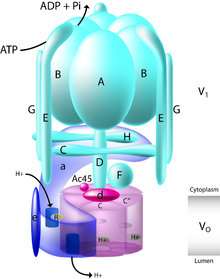
Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms.[1] V-ATPases acidify a wide array of intracellular organelles and pumps protons across the plasma membranes of numerous cell types. V-ATPases couple the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes of eukaryotic cells. It is generally seen as the polar opposite of ATP synthase because ATP synthase is a proton channel that uses the energy from a proton gradient to produce ATP. V-ATPase however, is a proton pump that uses the energy from ATP hydrolysis to produce a proton gradient.
The Archaea-type ATPase (A-ATPase) is a related group of ATPases found in archaea that often work as an ATP synthase. It forms a clade V/A-ATPase with V-ATPase. Most members of either group shuttle protons (H+
), but a few members have evolved to use sodium ions (Na+
) instead.
- ^ Nelson N, Perzov N, Cohen A, Hagai K, Padler V, Nelson H (January 2000). "The cellular biology of proton-motive force generation by V-ATPases". The Journal of Experimental Biology. 203 (Pt 1): 89–95. doi:10.1242/jeb.203.1.89. PMID 10600677.